QuantCenter is a multiple-module image analysis platform designed for whole-slide quantification in histopathology and molecular pathology. Thanks to its wide range of linkable modules and the ability to create their own image analysis algorithm, QuantCenter provides researchers with maximum flexibility in quantitative image analysis, contributing to world-class research results.
The latest available version of QuantCenter is 2.3. Please get in touch with us if you are interested!
QuantCenter is intended for use in tissue classification, IHC quantification and molecular pathology. It consists of ten individual modules that can be freely combined for world-class research results:
ScriptQuant: A unique tool for maximum flexibility
ScriptQuant is a brand-new image analysis module for QuantCenter. It provides a shell for user-created image analysis scripts in Python for maximum flexibility for the research user, enabling them to run their own image analysis algorithms within the QuantCenter framework.
Key Features
- Arbitrary script editor (VS Code by Microsoft is recommended)
- The user-created script can be saved into a scenario and handled like one
- Fast visual feedback: write your code and see the results immediately!
- ScriptQuant can be extended with external libraries (e.g. OpenCV, TensorFlow…)
- Well-documented API and use case examples
- ScriptQuant may provide a gateway for 3rd party AI developers
- Quantification results can be fed back into the 3DHISTECH infrastructure:
- Storage of quantification results (polygons, objects, regions, measured values)
- Visualization in SlideViewer as an overlay
- Statistics and Data visualization
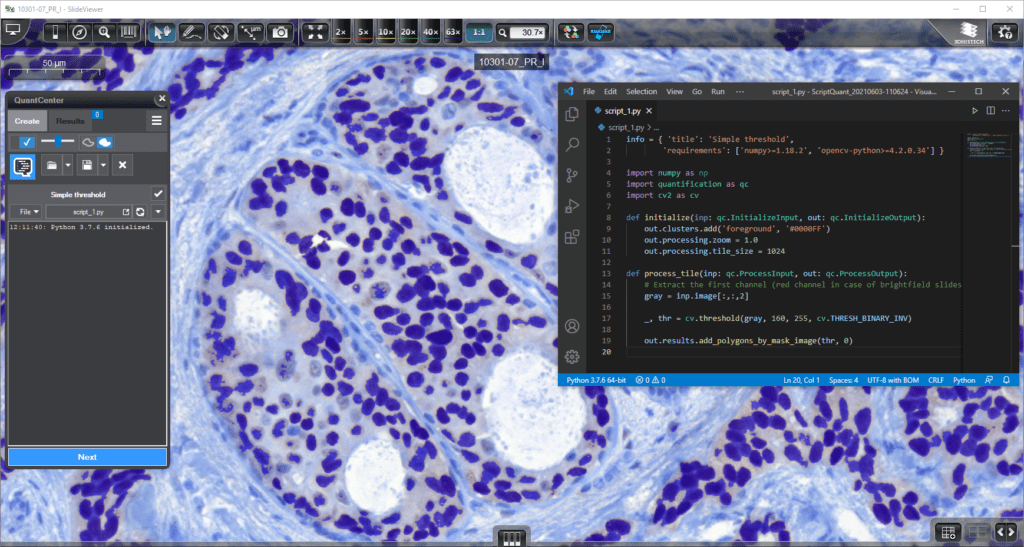
Image Analysis with QuantCenter
QuantCenter provides a range of image analysis modules that can be freely combined for best results.
Please also watch the available tutorials.
QuantCenter also offers an integrated data visualization mode and a batch analysis mode.
Tissue Classification
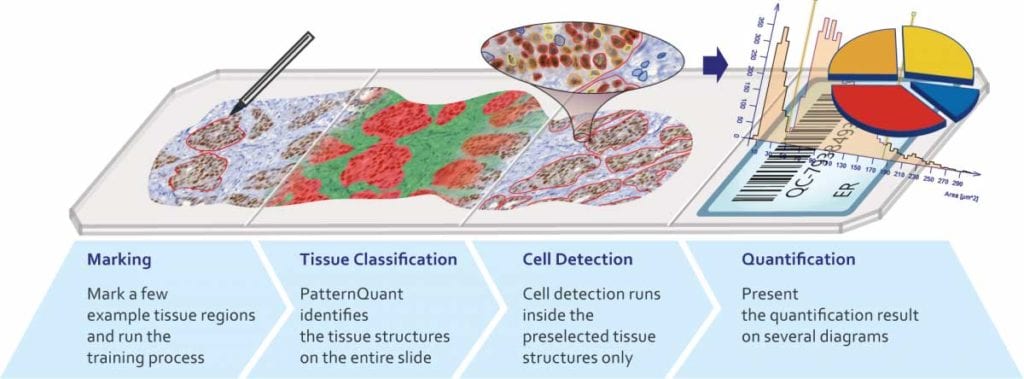
Color- and pattern-based image analysis solutions are used to identify different tissue elements in stained tissue. Instead of the time-consuming manual annotation, automated detection and annotation of a region of interest is available with a quick training on relevant tissue samples.
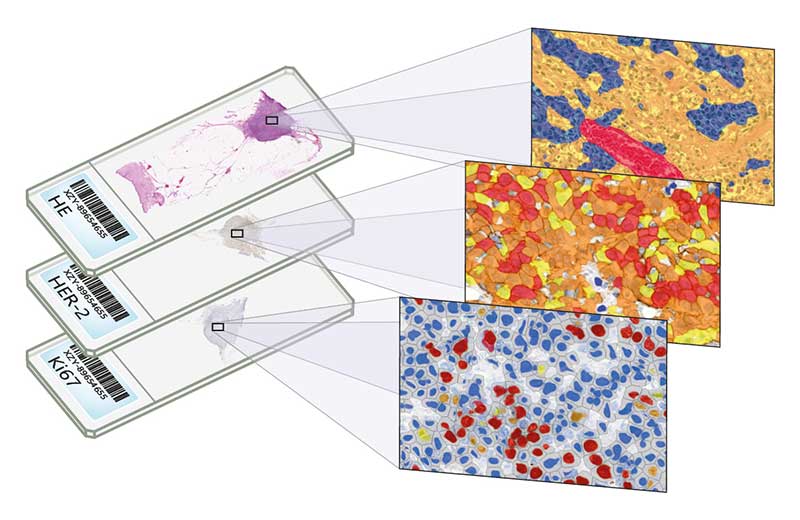
HistoQuant
HistoQuant is an image segmentation module which identifies stained tissue elements based on color and intensity features. This is an adequate solution for double-stain quantification or multiplex fluorescent analysis, with dual stain colocalization option. This module can analyze fluorescent and brightfield slides.
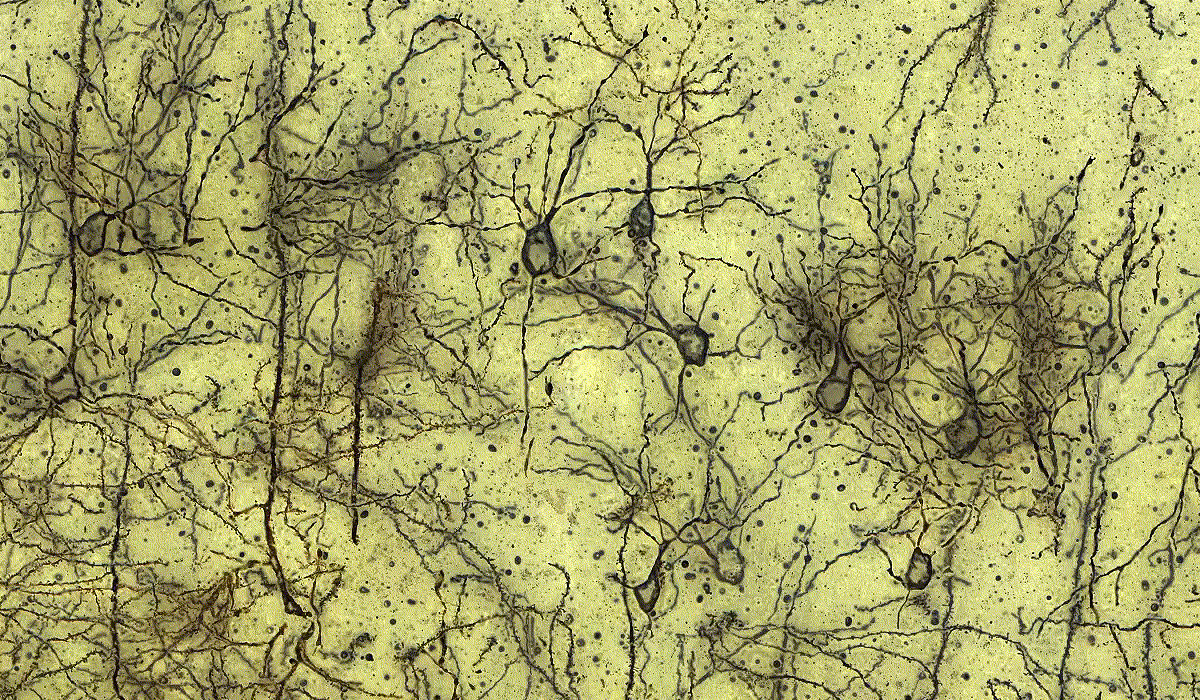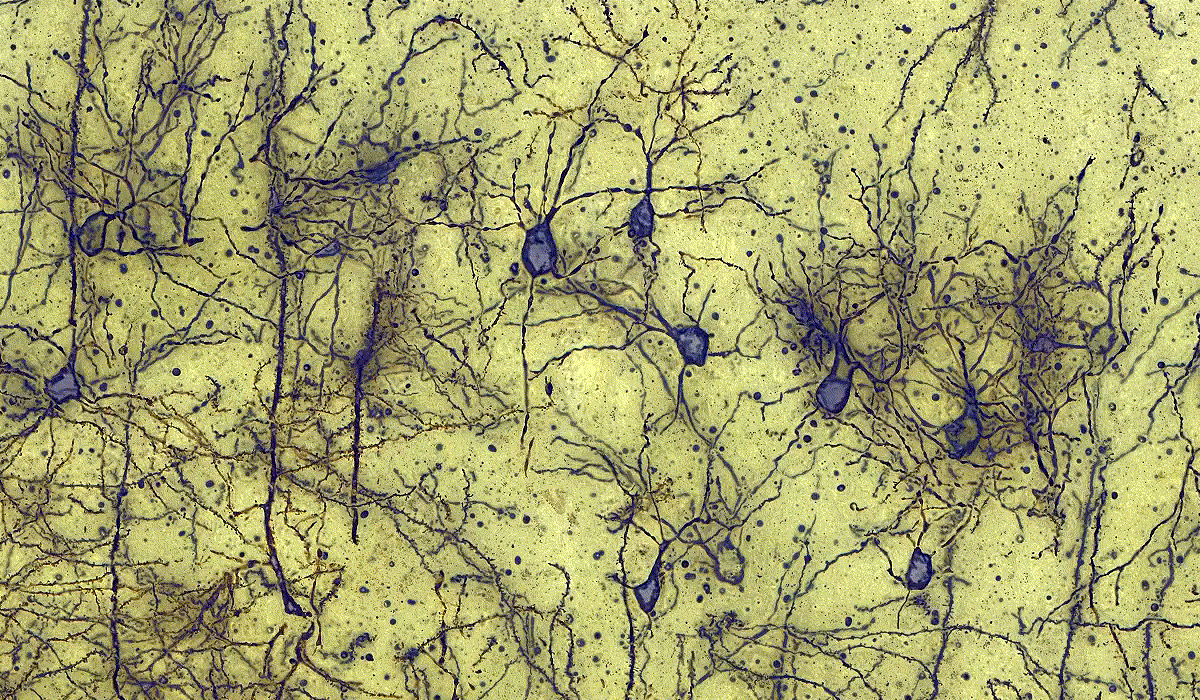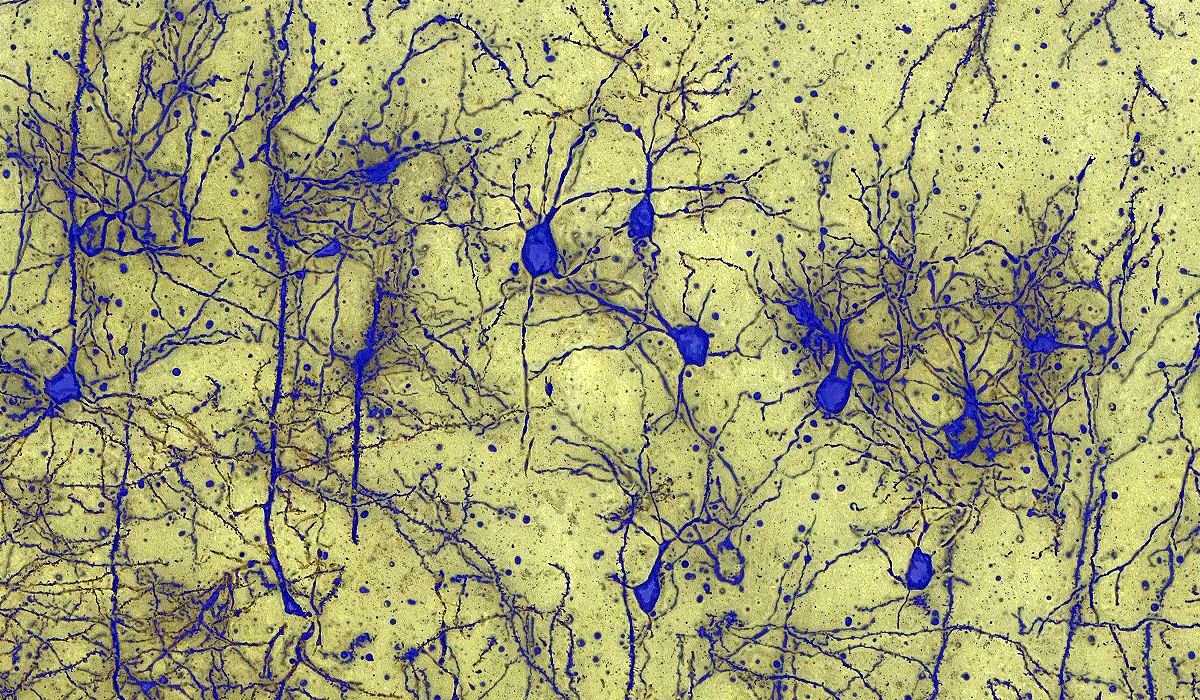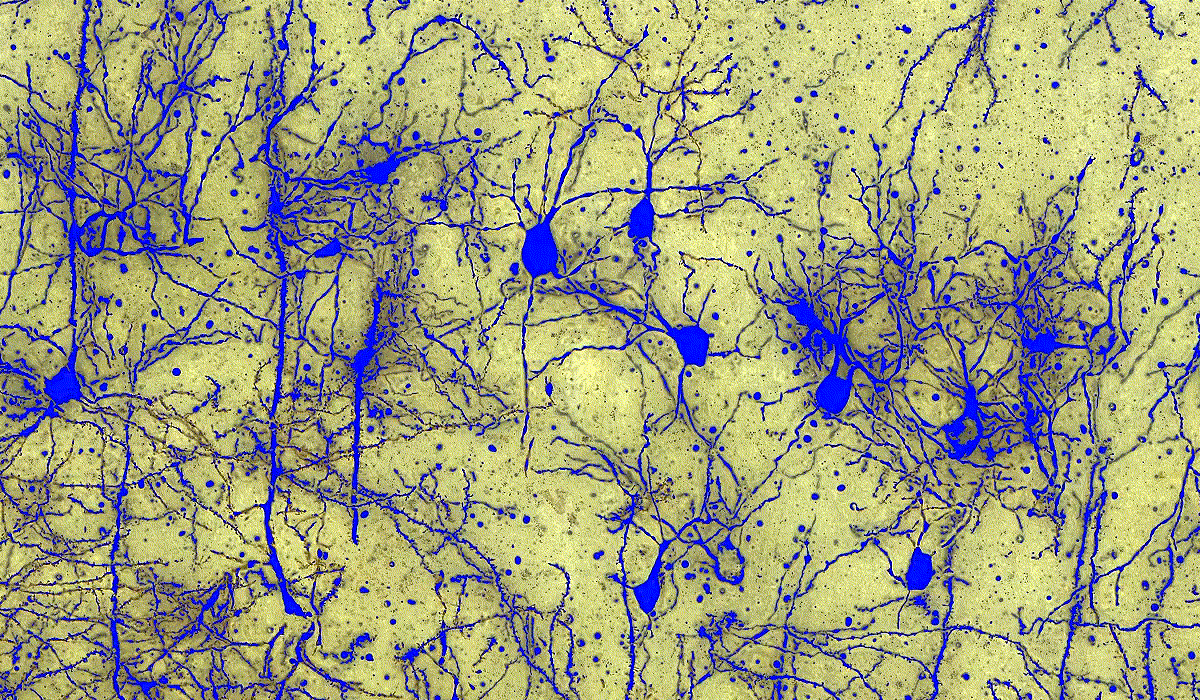
PatternQuant
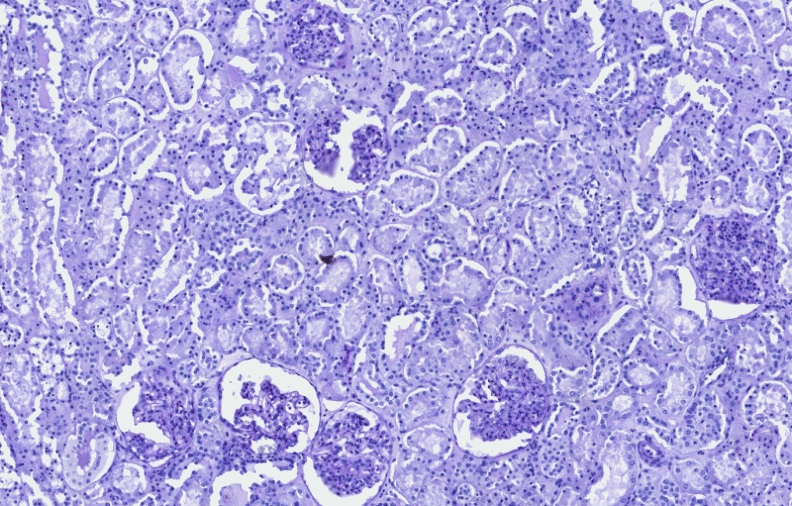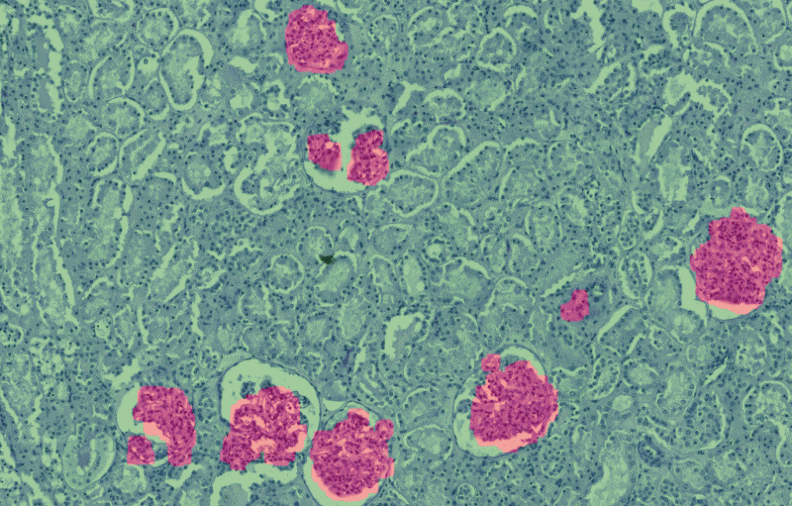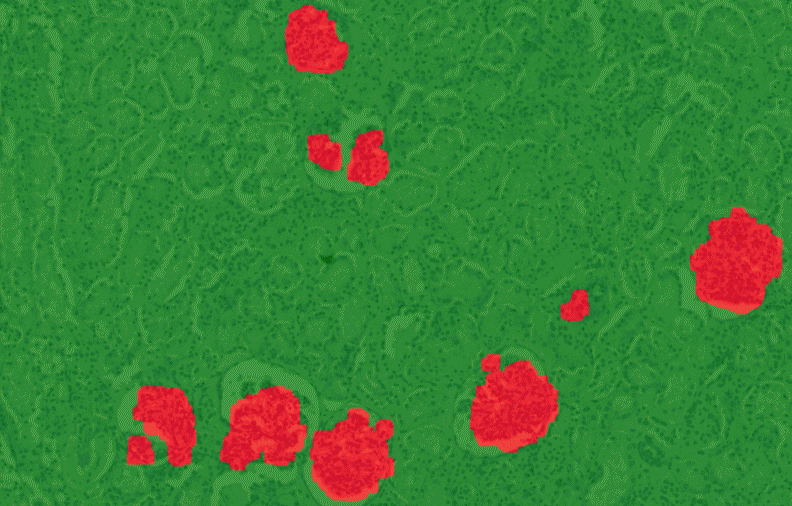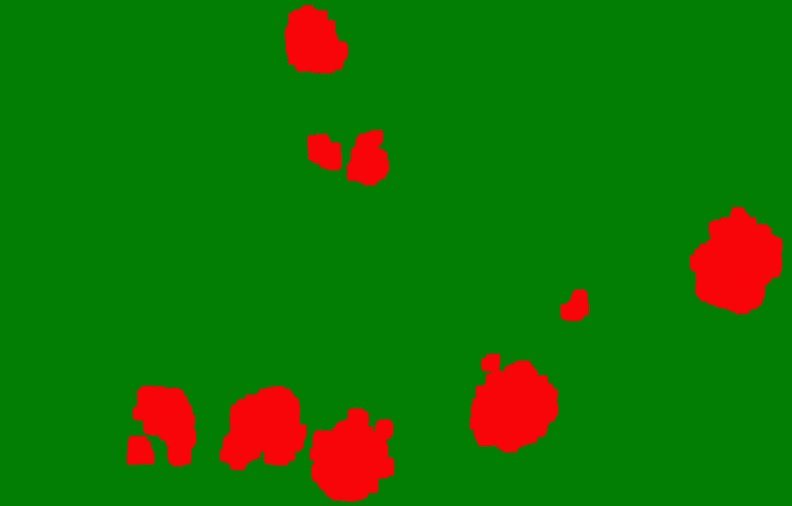
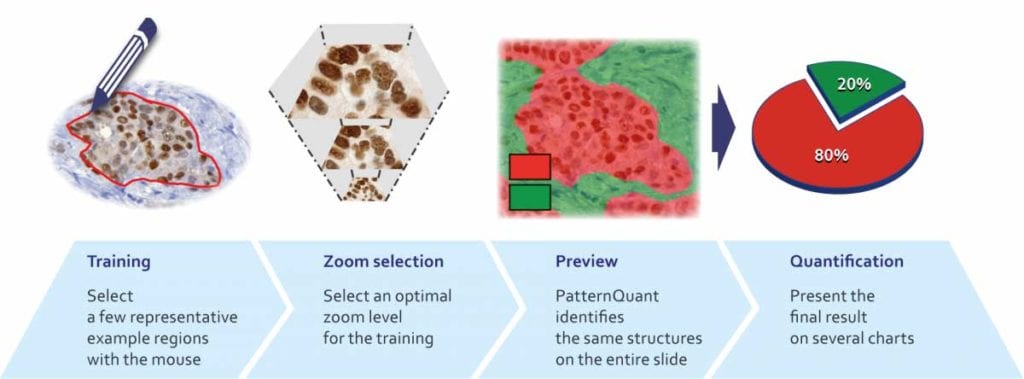
PatternQuant is a trainable pattern recognition module for tissue classification, tissue pre-segmentation and identification of several tissue structures. The artificial intelligence-based algorithm is able to learn and classify various tissue types based on their texture patterns and colour features. This module can analyze fluorescent and brightfield slides.
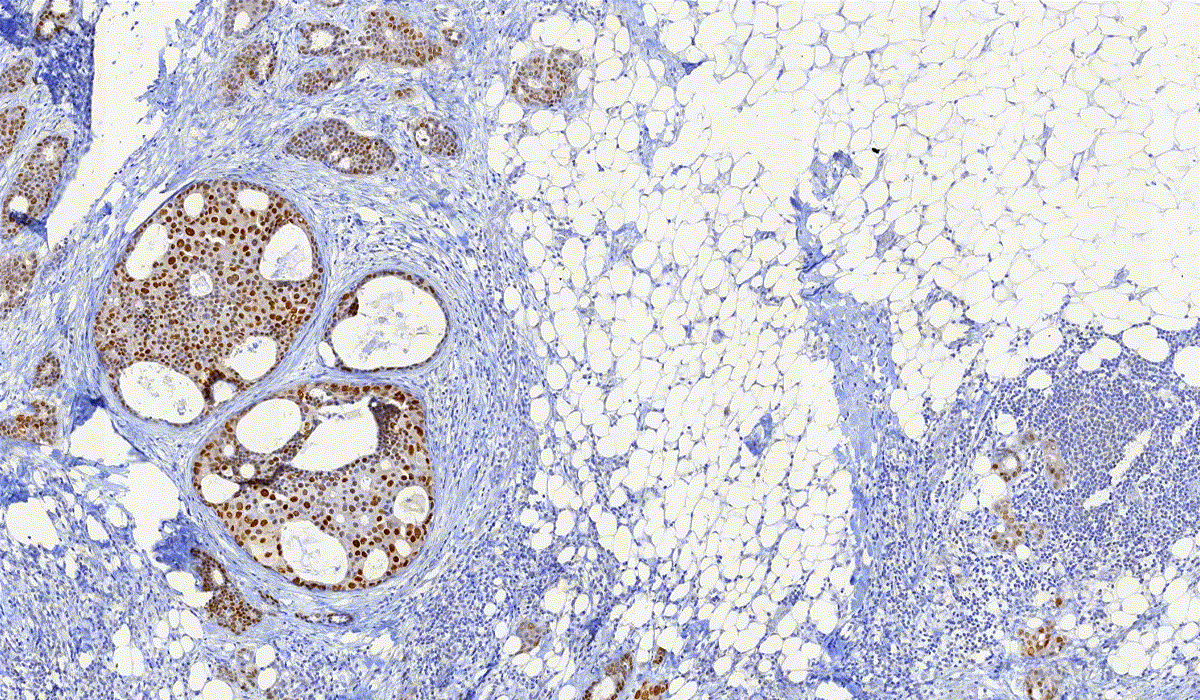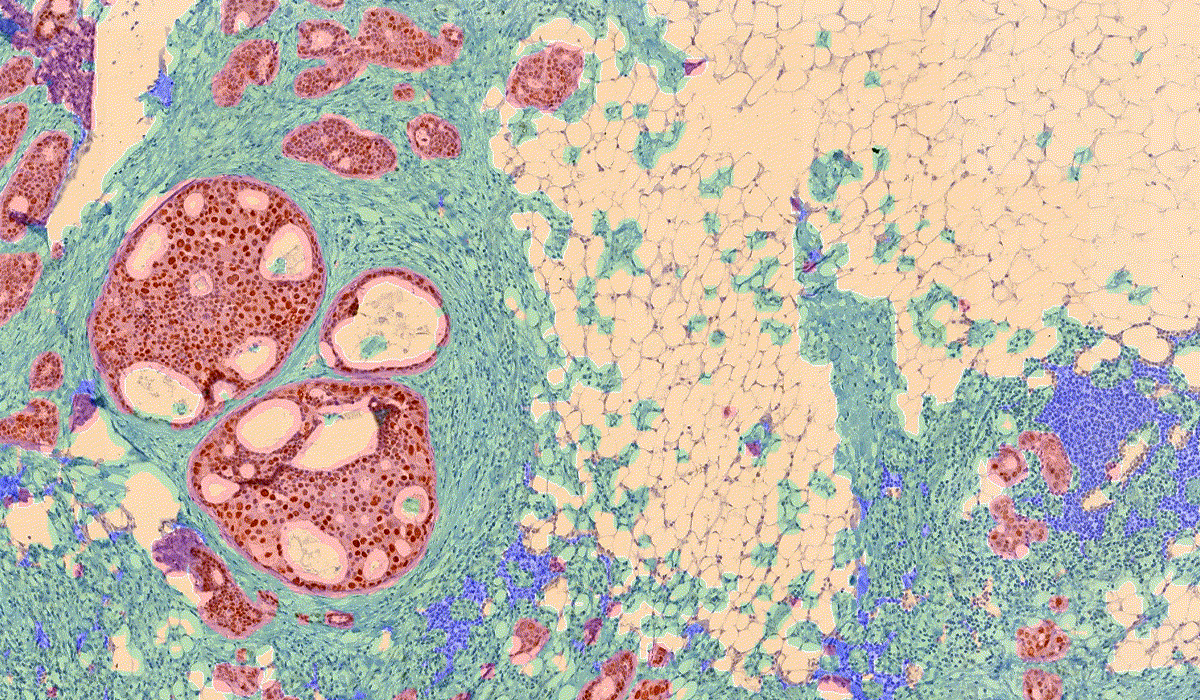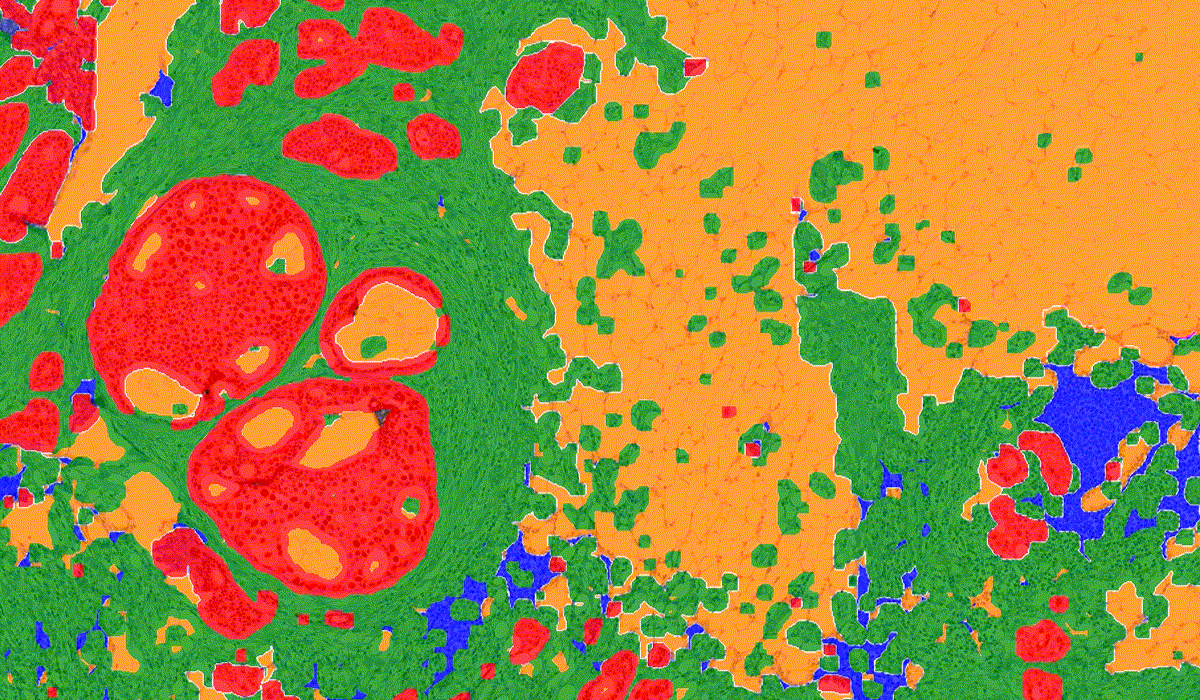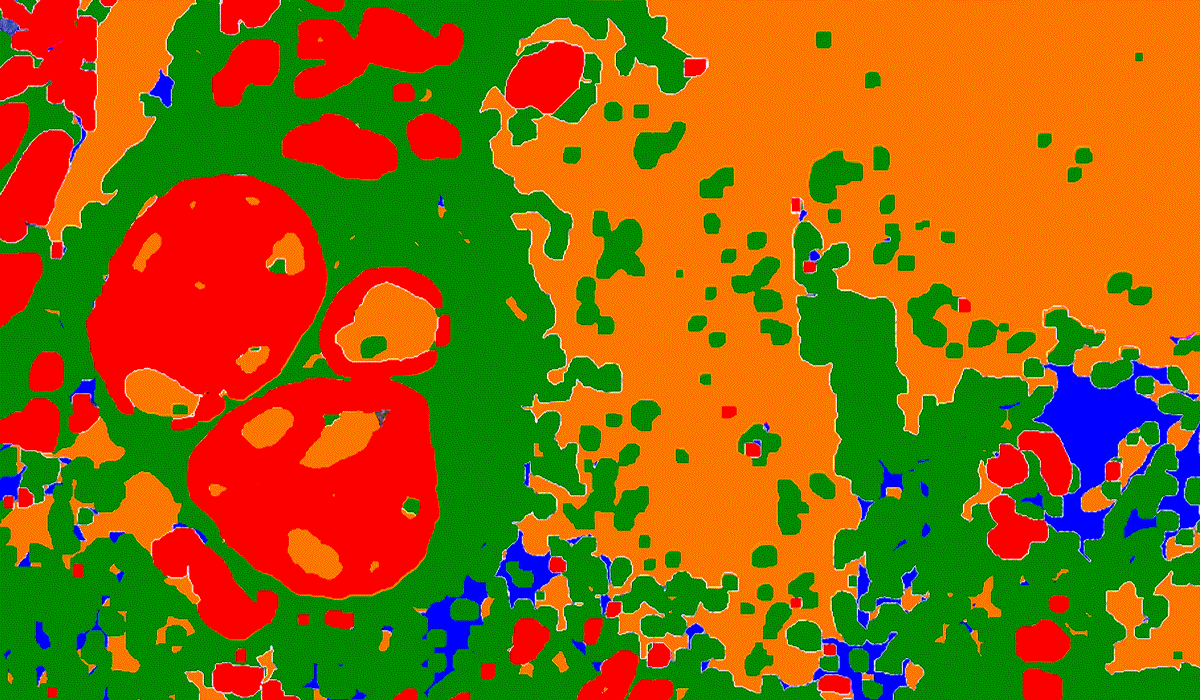
PatternQuant Plus
PatternQuant Plus is an improved alternative to PatternQuant for more challenging samples (low visual difference between the staining of tissue types within the sample) and for complex tissue segmentation projects when thorough training using deep learning on multiple slides is needed. This module can analyze fluorescent and brightfield slides.
IHC Quantification
A special IHC color calibration option is integrated into these quantification modules, facilitating software integration to the local laboratory. By applying this function the software can be calibrated to the sample quality (local laboratory stain protocol, or different stainer), resulting in an adequate measurement.
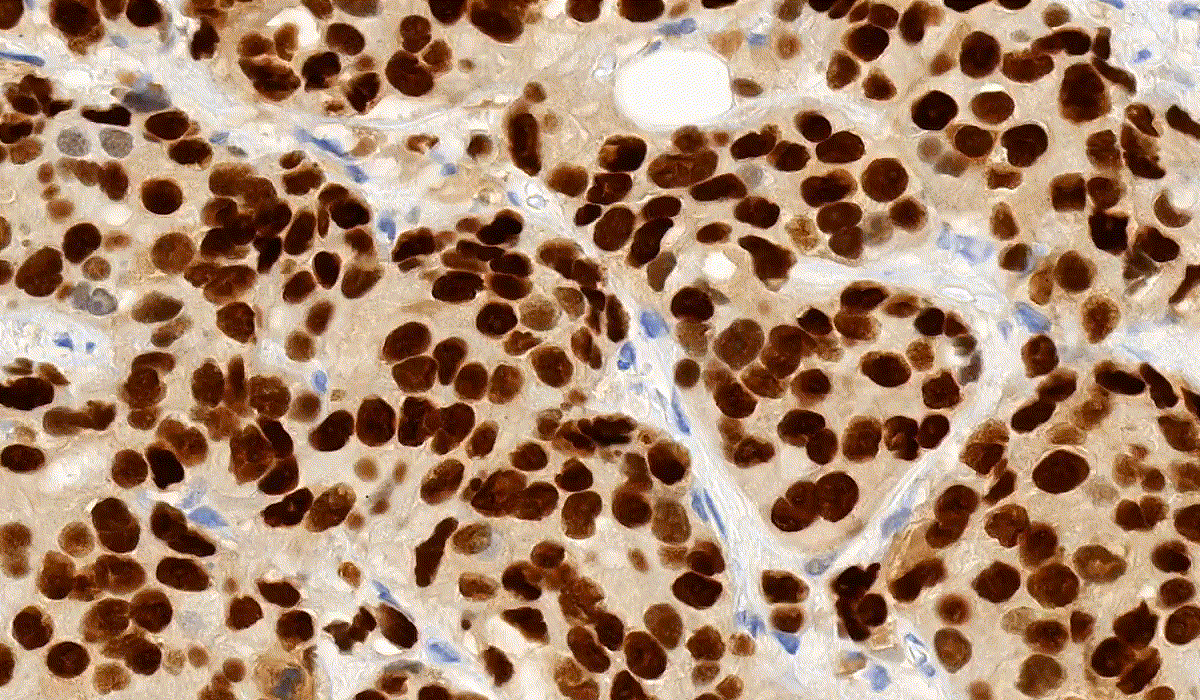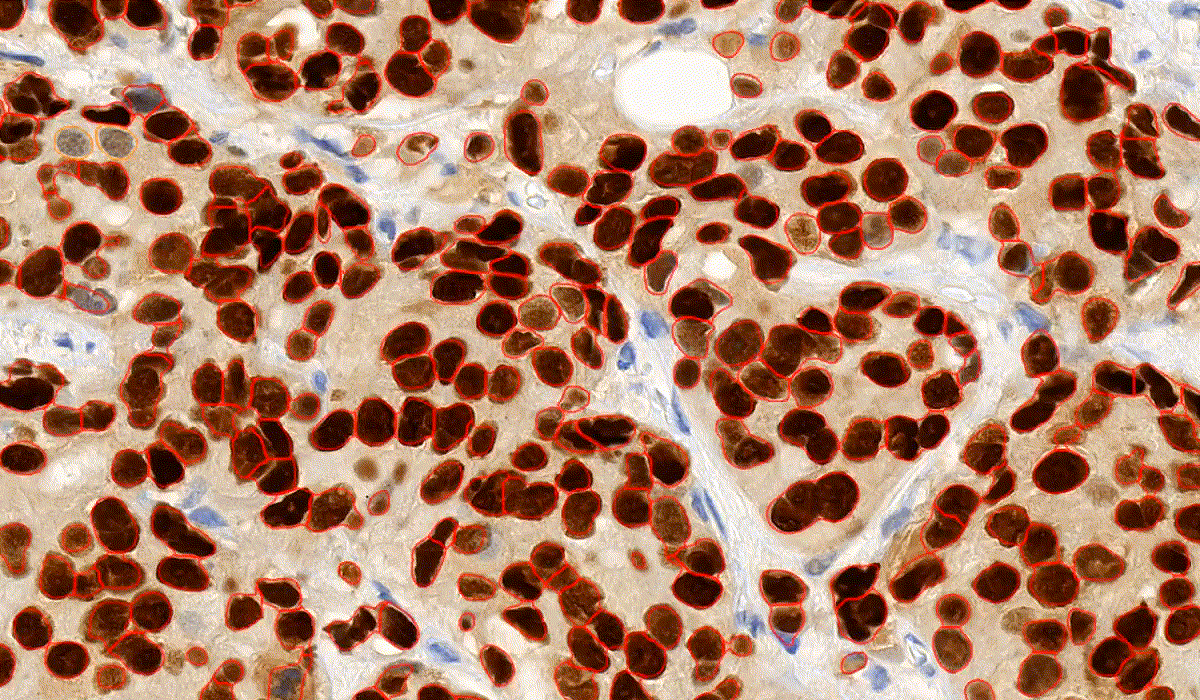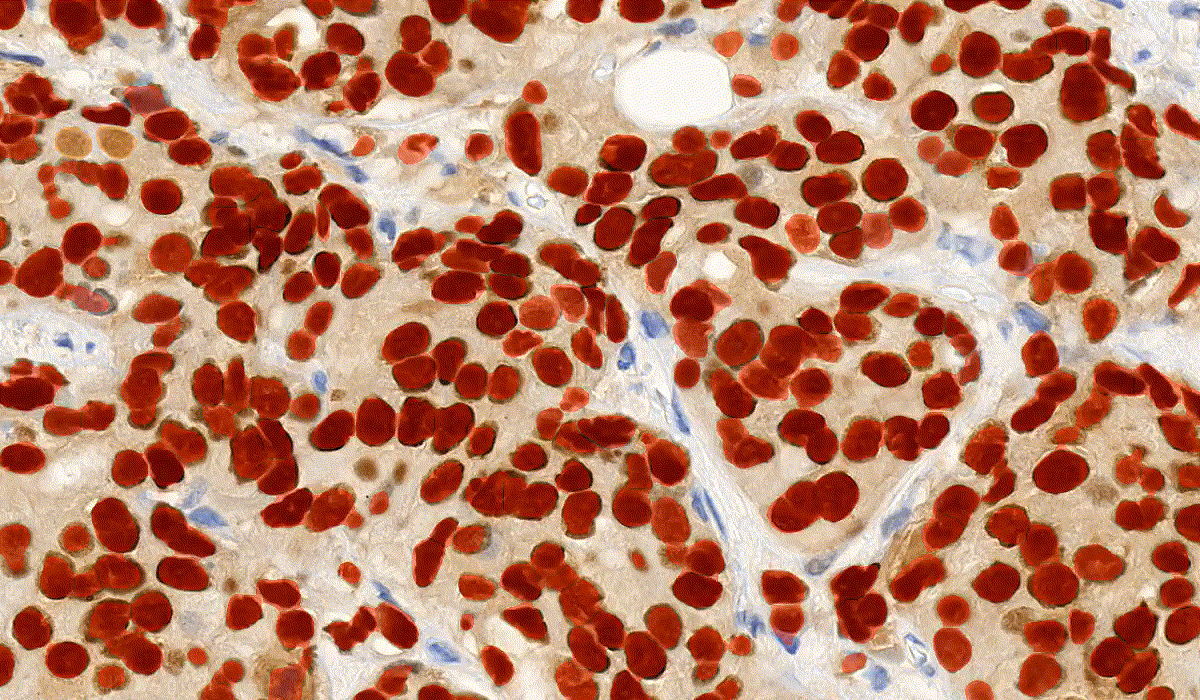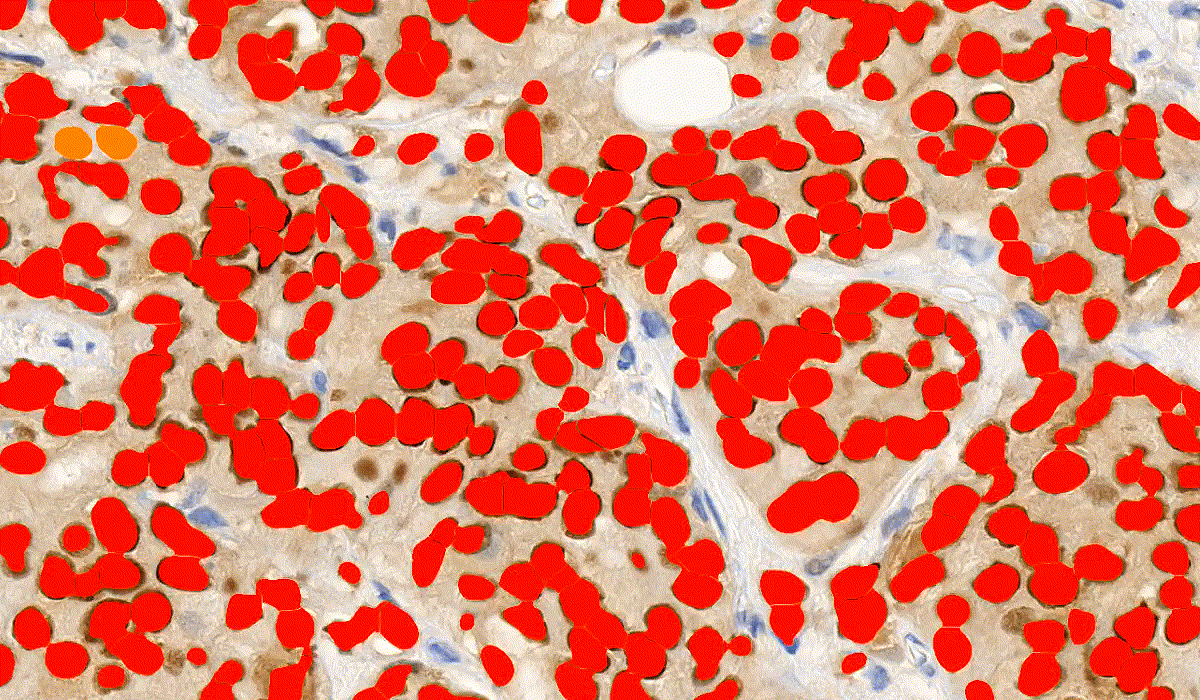
NuclearQuant
NuclearQuant is designed for cell nuclei detection and quantification of IHC stained samples. NuclearQuant measures several morphological features besides stain intensity. The cell nuclei classification and the final score are calculated by the intensity score and the proportion score.
This module has IVD approval for the analysis of Estrogen- and Progesterone-stained breast tissue samples.
MembraneQuant
MembraneQuant is a cell membrane detection software application that can be used for the quantification of IHC stained histological samples. This module measures cell morphology and stain density, reports intensity-based core ranges, overall scores and positivity percentages (including H-Score), distinguishing the continuous membrane stain from the moderate stain.
This module has IVD approval for the quantification of Her2 expression in breast tissue samples.
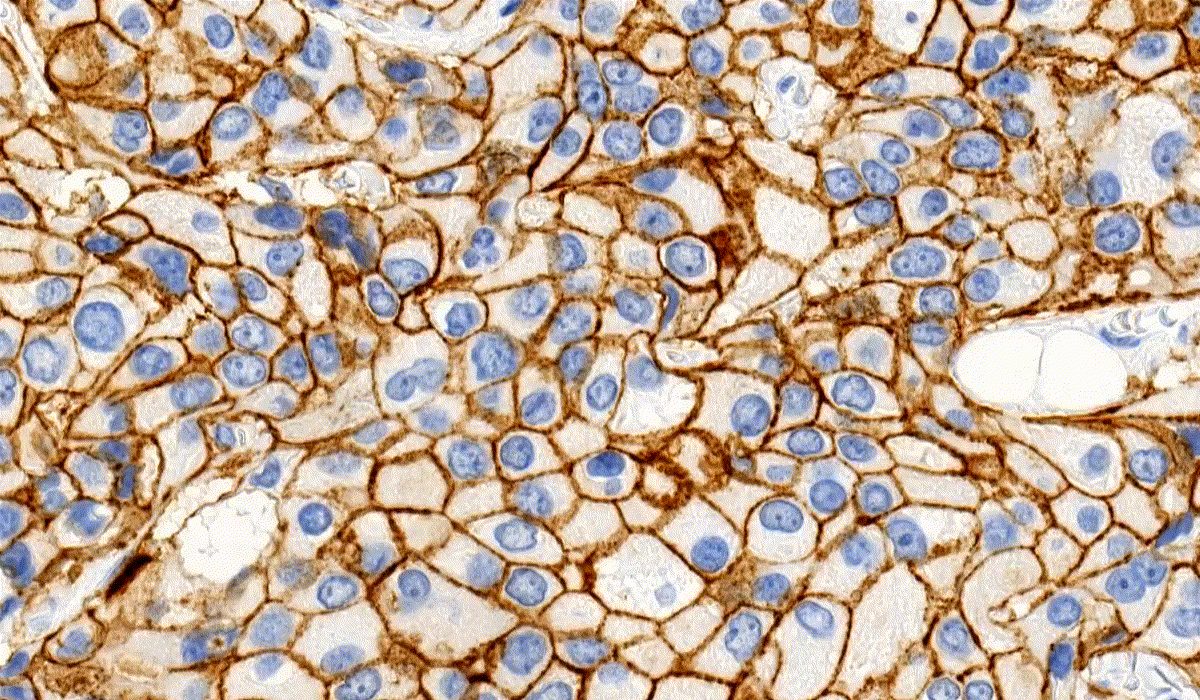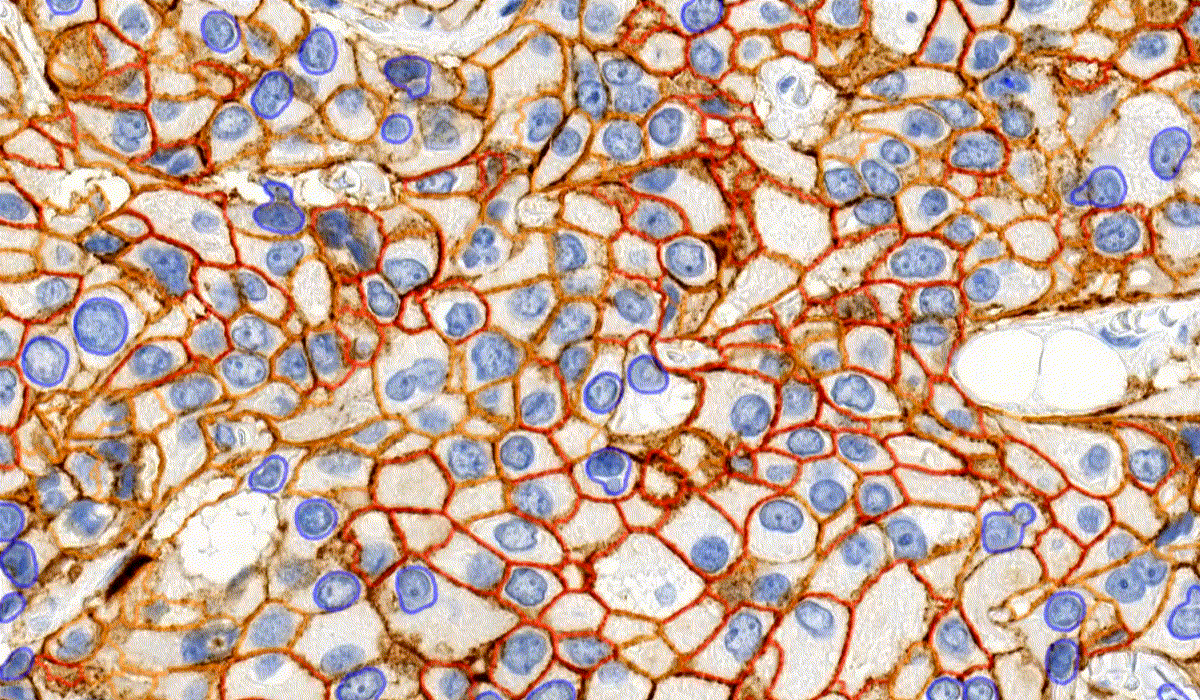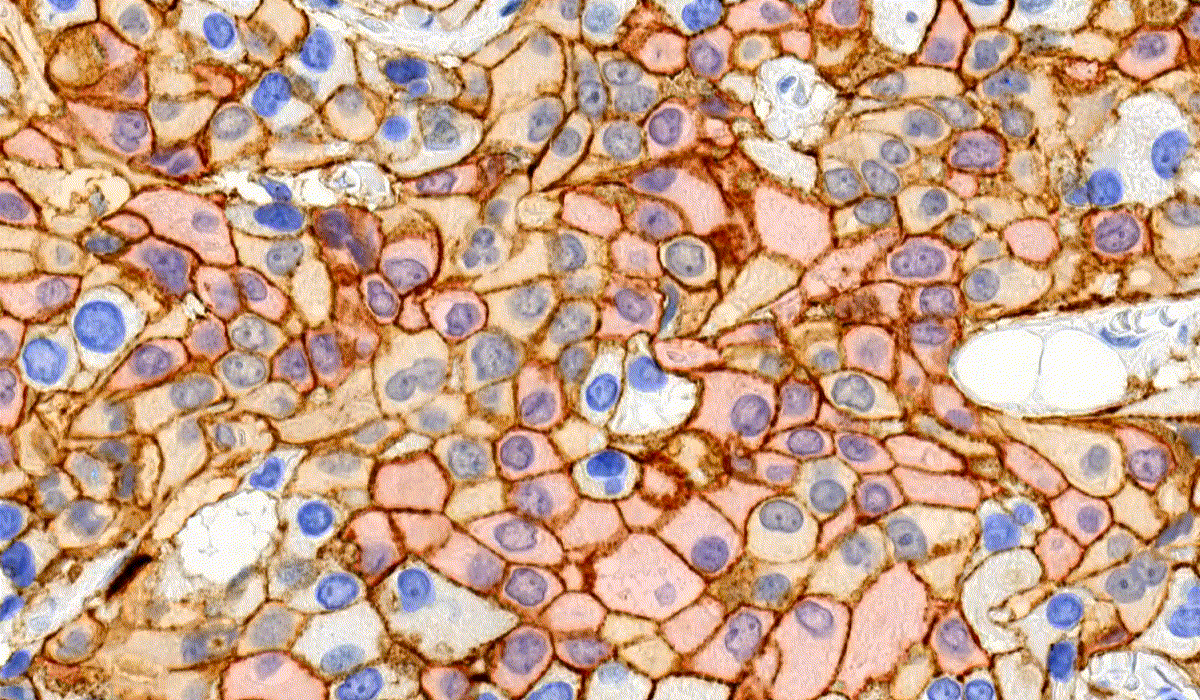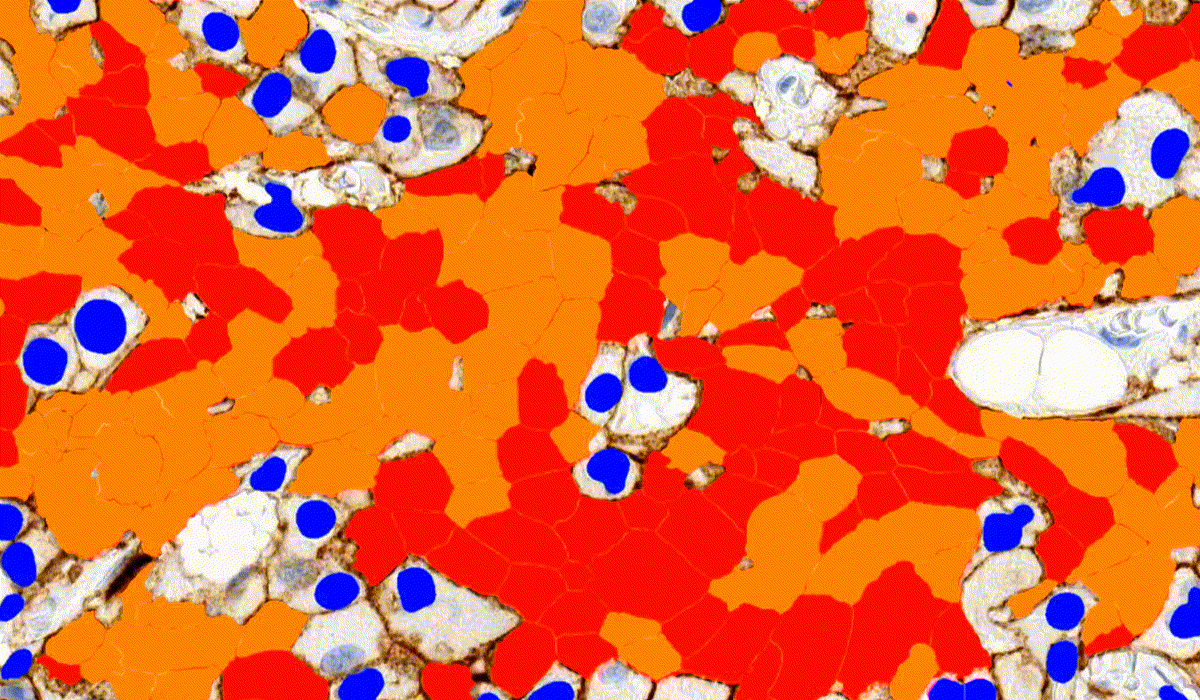
CellQuant
The Swiss army knife of 3DHISTECH’s image analysis tools, CellQuant is a versatile cell detection application, an optimal solution for investigating various IHC stainings (best suited for Ki67 slides). The application is adequate for cell nuclei, cytoplasmic and membrane marker quantification. The software reports the results like positivity ranges of cell nuclei, cytoplasm or membrane signals. This module can analyze fluorescent and brightfield slides.
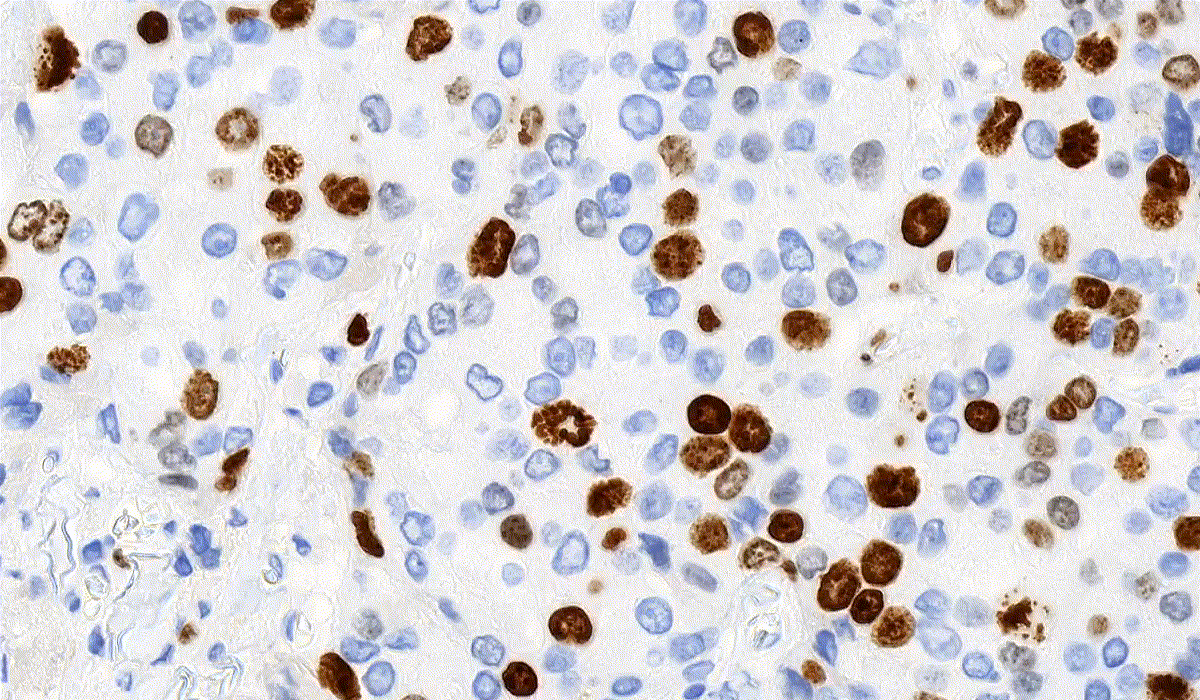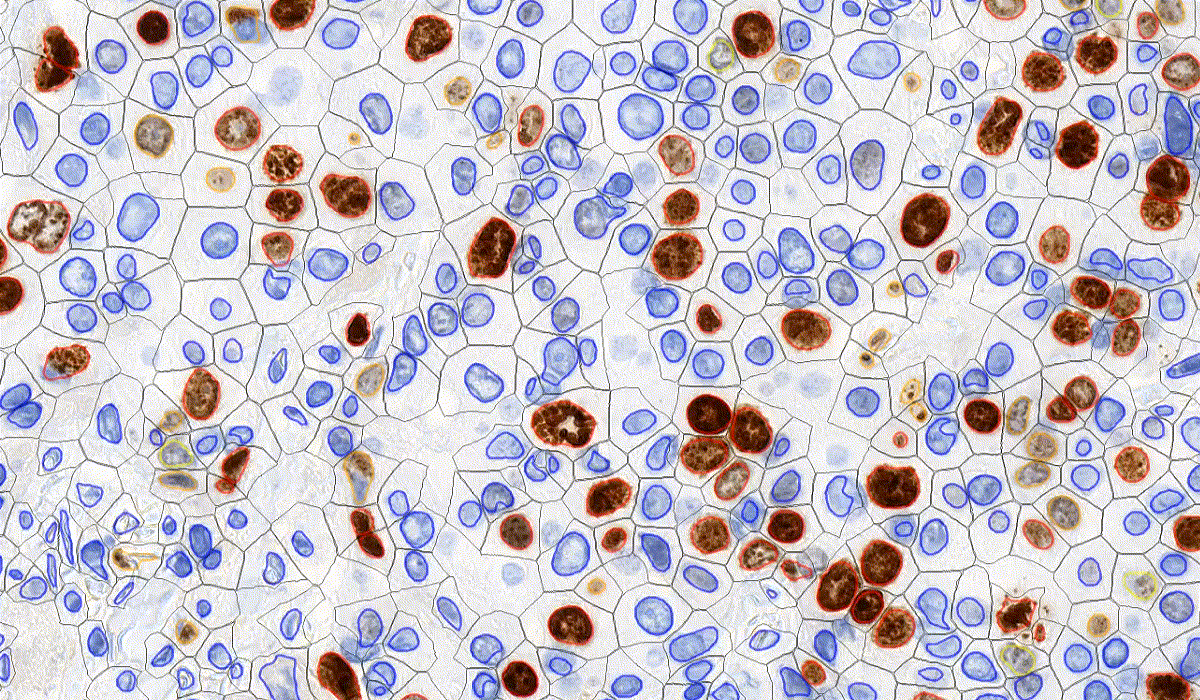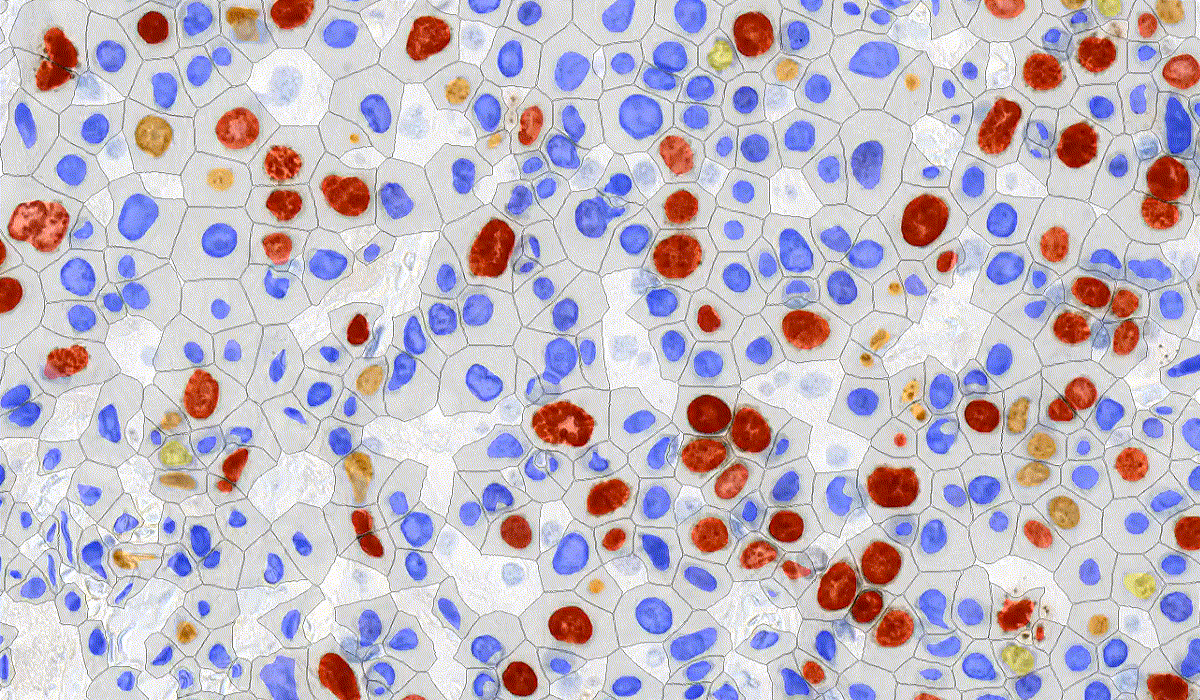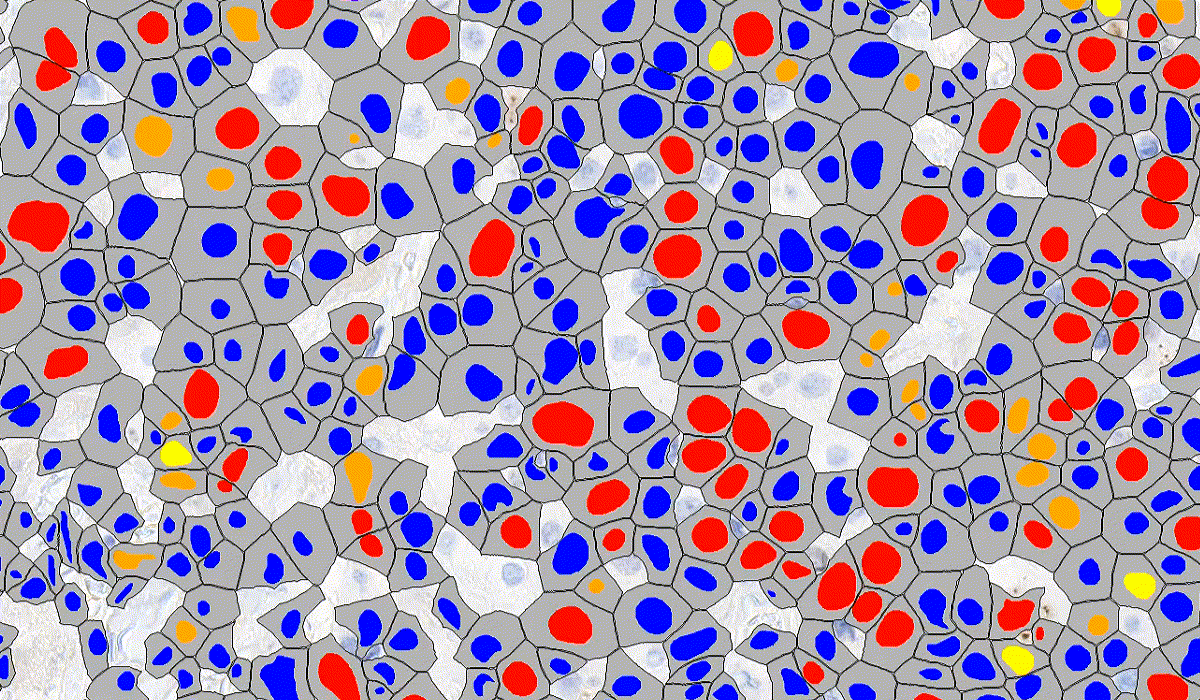
DensitoQuant
DensitoQuant is an easy-to-use, fast and accurate, stain-intensity-based IHC quantification tool. The application identifies the positive stain, based on an automatic color separation method through which individual positive pixels are counted and classified based on intensity and threshold ranges. This way, an H-Score can be calculated based on the proportion of positive and negative pixels.
Molecular Pathology
Fluorescent in situ hybridization (FISH) and Chromogenic in situ hybridization (CISH) are routinely used methods in histopathological diagnostics. The visual evaluation of these samples is a difficult task, and typically a time-consuming process and prone to human errors that can be eliminated by using image analysis solutions.
FISHQuant
FISHQuant is a powerful cancer and cytogenetic application dedicated to quantifying FISH (Fluorescent In Situ Hybridization) signals on tissue samples of solid tumour diseases like breast cancer, lung cancer, sarcoma symptoms, and lymphomas. This module is suitable for examining genetic mutations (gene amplification, deletion) and chromosomal aberrations (polysomy, aneusomy) on FISH slides. This module has IVD approval.
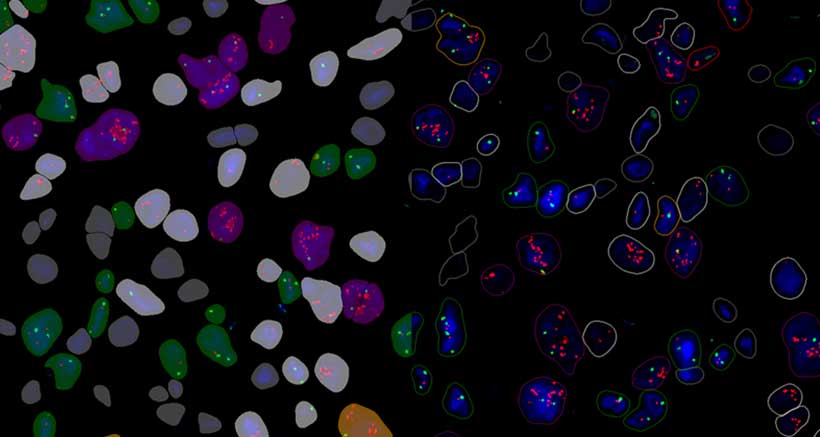
CISHQuant
CISHQuant quantifies CISH (Chromogenic In Situ Hybridization) stained samples. The algorithm can be calibrated to the staining protocol and quality by using an integrated colour setting tool. This module is suitable for examining genetic mutations (gene amplification, deletion) and chromosomal aberrations (polysomy, aneusomy) on CISH slides.
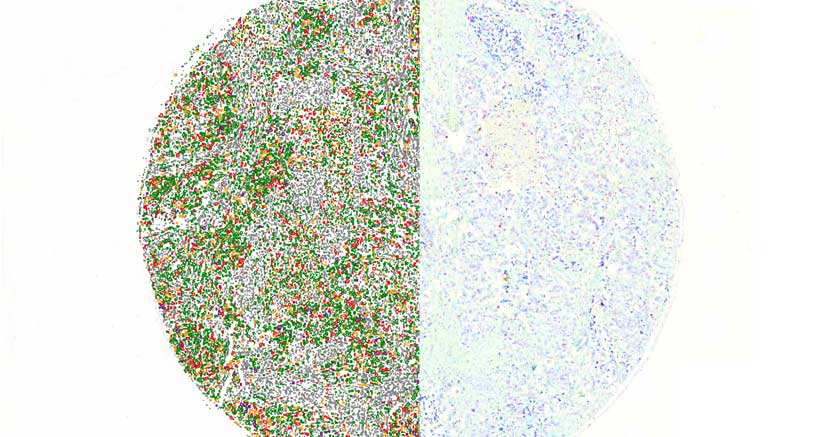
Batch Analysis
Using the batch analysis mode, multiple digital slides can be examined in the background, saving a significant amount of time. Measurements can be run after slides, annotations and profiles have been selected. All measurement results can be exported into an Excel file.
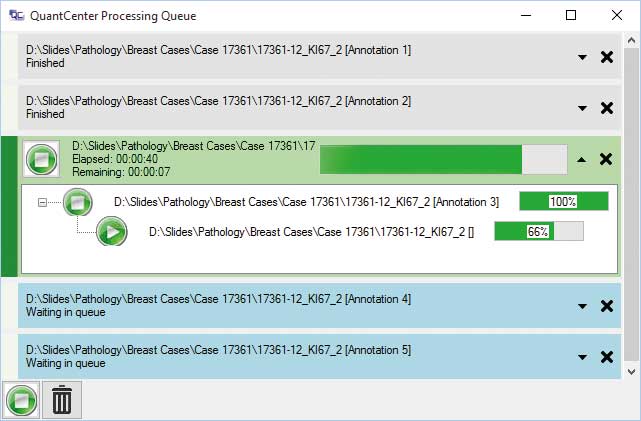
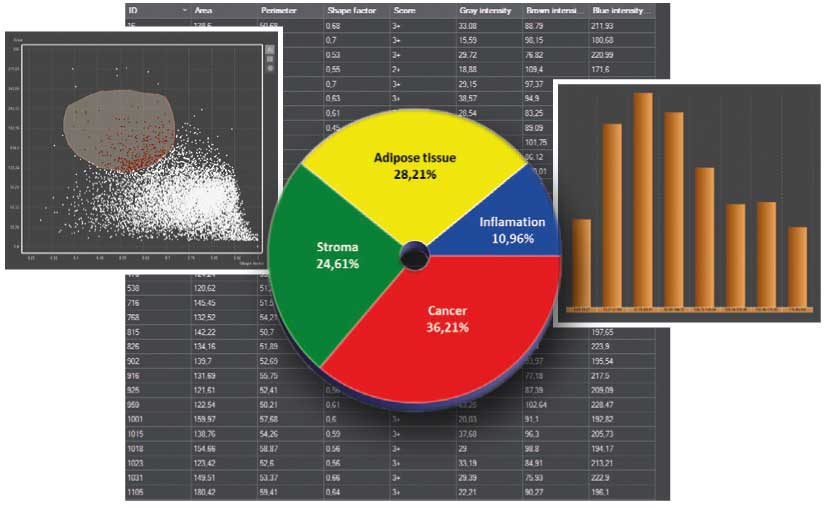
Data Visualization (DVT)
An integrated data visualization mode is available for each module. The result can be visualized on a table, scatterplot, pie chart or a histogram by using this feature. There are integrated galleries where the detected and classified cells can be presented.
Tutorials
Her2stain quantification using PatternQuant and MembraneQuant
Breast Cancer Tissue Analysis using PatternQuant and NuclearQuant

